Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: May 2010
- TU Wien Library:
- Diploma Examination: 11. May 2010
- First Supervisor:
Abstract
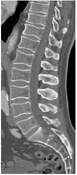
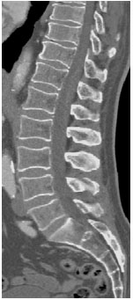
Medical Image Processing is a growing field in medicine and plays an important role in medical decision making. Computer-based segmentation of anatomies in data made by imaging modalities supports clinicians and speeds up their diagnosis making compared to doing it manually. Computed Tomography (CT) is an imaging modality for slice-wise three dimensional reconstruction of the human body in the form of volumetric data which is especially applicable for imaging of bony structures and so for the vertebral column. Most bony structures, such as vertebrae, are characterised by complex shape and texture appearances which turns its segmentation into a difficult task. Model-based segmentation approaches are promising techniques to cope with variations in form and texture of the anatomy of interest. This is done by incorporating information about shape and texture appearance gained from an imaging modality in a model. The model can then be applied to segment the object of interest in target data, however most of the model-based approaches need a model intialisation for a fast and reliable segmentation of the object of interest. This thesis was motivated by novel works on fast anatomical structure localisation with Markov Random Fields (MRFs) and focuses on the sparse structure localisation of single vertebrae in CT scans for a subsequent model initialisation of more sophisticated segmentation algorithms. A MRF based model of appearance, which employs local information in regions around anatomical landmarks and geometrical information through connections between adjacent landmarks, is built on volumetric CT datasets of lumbar vertebrae. The MRF based model is built on a 6 landmark configuration in vertebra volumetric data and is additionally matched with target data. This is done by finding a best fit MRF matching by the Max-sum algorithm among feature points found by a decision tree based feature detection algorithm called probabilistic boosting tree (PBT). Anatomical landmark regions are described by vector spin-images and shape index histograms. Adjacency information is extracted by Delaunay tetrahedralisation where distances and gradient-related angles describe connections between adjacent regions. The results on single lumbar vertebra CT scans show that the MRF approach is applicable on volumetric CT datasets with an accuracy enough for supporting more sophisticated segmentation algorithms such as Active Appearance Models (AAMs).Additional Files and Images
Weblinks
No further information available.BibTeX
@mastersthesis{major-2010-mrf,
title = "Markov Random Field Based Structure Localisation of
Vertebrae for 3D-Segmentation of the Spine in CT Volume Data",
author = "David Major",
year = "2010",
abstract = "Medical Image Processing is a growing field in medicine and
plays an important role in medical decision making.
Computer-based segmentation of anatomies in data made by
imaging modalities supports clinicians and speeds up their
diagnosis making compared to doing it manually. Computed
Tomography (CT) is an imaging modality for slice-wise three
dimensional reconstruction of the human body in the form of
volumetric data which is especially applicable for imaging
of bony structures and so for the vertebral column. Most
bony structures, such as vertebrae, are characterised by
complex shape and texture appearances which turns its
segmentation into a difficult task. Model-based segmentation
approaches are promising techniques to cope with variations
in form and texture of the anatomy of interest. This is done
by incorporating information about shape and texture
appearance gained from an imaging modality in a model. The
model can then be applied to segment the object of interest
in target data, however most of the model-based approaches
need a model intialisation for a fast and reliable
segmentation of the object of interest. This thesis was
motivated by novel works on fast anatomical structure
localisation with Markov Random Fields (MRFs) and focuses on
the sparse structure localisation of single vertebrae in CT
scans for a subsequent model initialisation of more
sophisticated segmentation algorithms. A MRF based model of
appearance, which employs local information in regions
around anatomical landmarks and geometrical information
through connections between adjacent landmarks, is built on
volumetric CT datasets of lumbar vertebrae. The MRF based
model is built on a 6 landmark configuration in vertebra
volumetric data and is additionally matched with target
data. This is done by finding a best fit MRF matching by the
Max-sum algorithm among feature points found by a decision
tree based feature detection algorithm called probabilistic
boosting tree (PBT). Anatomical landmark regions are
described by vector spin-images and shape index histograms.
Adjacency information is extracted by Delaunay
tetrahedralisation where distances and gradient-related
angles describe connections between adjacent regions. The
results on single lumbar vertebra CT scans show that the MRF
approach is applicable on volumetric CT datasets with an
accuracy enough for supporting more sophisticated
segmentation algorithms such as Active Appearance Models
(AAMs).",
month = may,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2010/major-2010-mrf/",
}
 image
image paper
paper