Information
- Publication Type: Journal Paper (without talk)
- Workgroup(s)/Project(s):
- Date: May 2014
- ISSN: 0167-7055
- Journal: Computer Graphics Forum
- Number: 6
- Volume: 33
- Pages: 276 – 287
- Keywords: clustering, implicit surfaces, level of detail algorithms, scientific visualization, Computer Applications
Abstract
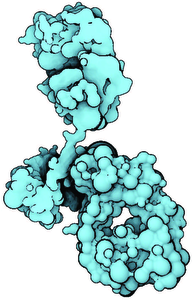
Molecular visualization is often challenged with rendering of large molecular structures in real time. We introduce a novel approach that enables us to show even large protein complexes. Our method is based on the level-of-detail concept, where we exploit three different abstractions combined in one visualization. Firstly, molecular surface abstraction exploits three different surfaces, solvent-excluded surface (SES), Gaussian kernels and van der Waals spheres, combined as one surface by linear interpolation. Secondly, we introduce three shading abstraction levels and a method for creating seamless transitions between these representations. The SES representation with full shading and added contours stands in focus while on the other side a sphere representation of a cluster of atoms with constant shading and without contours provide the context. Thirdly, we propose a hierarchical abstraction based on a set of clusters formed on molecular atoms. All three abstraction models are driven by one importance function classifying the scene into the near-, mid- and far-field. Moreover, we introduce a methodology to render the entire molecule directly using the A-buffer technique, which further improves the performance. The rendering performance is evaluated on series of molecules of varying atom counts.Additional Files and Images
Weblinks
BibTeX
@article{Viola_Ivan_CLD,
title = "Continuous Levels-of-Detail and Visual Abstraction for
Seamless Molecular Visualization",
author = "Julius Parulek and Daniel J\"{o}nsson and Timo Ropinski and
Stefan Bruckner and Anders Ynnerman and Ivan Viola",
year = "2014",
abstract = "Molecular visualization is often challenged with rendering
of large molecular structures in real time. We introduce a
novel approach that enables us to show even large protein
complexes. Our method is based on the level-of-detail
concept, where we exploit three different abstractions
combined in one visualization. Firstly, molecular surface
abstraction exploits three different surfaces,
solvent-excluded surface (SES), Gaussian kernels and van der
Waals spheres, combined as one surface by linear
interpolation. Secondly, we introduce three shading
abstraction levels and a method for creating seamless
transitions between these representations. The SES
representation with full shading and added contours stands
in focus while on the other side a sphere representation of
a cluster of atoms with constant shading and without
contours provide the context. Thirdly, we propose a
hierarchical abstraction based on a set of clusters formed
on molecular atoms. All three abstraction models are driven
by one importance function classifying the scene into the
near-, mid- and far-field. Moreover, we introduce a
methodology to render the entire molecule directly using the
A-buffer technique, which further improves the performance.
The rendering performance is evaluated on series of
molecules of varying atom counts.",
month = may,
issn = "0167-7055",
journal = "Computer Graphics Forum",
number = "6",
volume = "33",
pages = "276--287",
keywords = "clustering, implicit surfaces, level of detail algorithms,
scientific visualization, Computer Applications",
URL = "https://www.cg.tuwien.ac.at/research/publications/2014/Viola_Ivan_CLD/",
}