Information
- Publication Type: PhD-Thesis
- Workgroup(s)/Project(s):
- Date: October 2016
- Date (Start): 2013
- Date (End): 2016
- TU Wien Library:
- 1st Reviewer: Ivan Viola

- 2nd Reviewer: C. Hansen
- Rigorosum: 23. November 2016
- First Supervisor: Ivan Viola

Abstract
Macromolecules, such as proteins, are the building blocks of the machinery of life, and therefore are essential to the comprehension of physiological processes. In physiology, illustrations and animations are often utilized as a mean of communication because they can easily be understood with little background knowledge. However, their realization
requires numerous months of manual work, which is both expensive and time consuming.
Computational biology experts produce everyday large amount of data that is publicly available and that contains valuable information about the structure and also the function of these macromolecules. Instead of relying on manual work to generate illustrative
visualizations of the cell biology, we envision a solution that would utilize all the data already available in order to streamline the creation process.
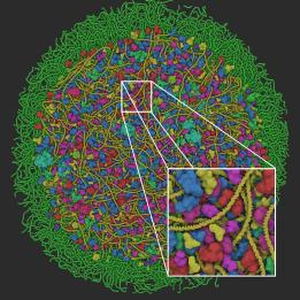
In this thesis are presented several contributions that aim at enabling our vision. First, a novel GPU-based rendering pipeline that allows interactive visualization of realistic molecular datasets comprising up to hundreds of millions of macromolecules. The rendering pipeline is embedded into a popular game engine and well known computer graphics optimizations were adapted to support this type of data, such as level-of-detail, instancing and occlusion queries. Secondly, a new method for authoring cutaway views and improving spatial exploration of crowded molecular landscapes. The system relies on the use of clipping objects that are manually placed in the scene and on visibility
equalizers that allows fine tuning of the visibility of each species present in the scene.
Agent-based modeling produces trajectory data that can also be combined with structural information in order to animate these landscapes. The snapshots of the trajectories are often played in fast-forward to shorten the length of the visualized sequences, which also renders potentially interesting events occurring at a higher temporal resolution invisible. The third contribution is a solution to visualize time-lapse of agent-based
simulations that also reveals hidden information that is only observable at higher temporal resolutions. And finally, a new type of particle-system that utilize quantitative models as input and generate missing spatial information to enable the visualization of molecular trajectories and interactions. The particle-system produces a similar visual output as
traditional agent-based modeling tools for a much lower computational footprint and
allows interactive changing of the simulation parameters, which was not achievable with previous methods.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
No further information available.
BibTeX
@phdthesis{LeMuzic_2016_PhD,
title = "From Atoms to Cells: Interactive and Illustrative
Visualization of Digitally Reproduced Lifeforms",
author = "Mathieu Le Muzic",
year = "2016",
abstract = "Macromolecules, such as proteins, are the building blocks of
the machinery of life, and therefore are essential to the
comprehension of physiological processes. In physiology,
illustrations and animations are often utilized as a mean of
communication because they can easily be understood with
little background knowledge. However, their realization
requires numerous months of manual work, which is both
expensive and time consuming. Computational biology experts
produce everyday large amount of data that is publicly
available and that contains valuable information about the
structure and also the function of these macromolecules.
Instead of relying on manual work to generate illustrative
visualizations of the cell biology, we envision a solution
that would utilize all the data already available in order
to streamline the creation process. In this thesis are
presented several contributions that aim at enabling our
vision. First, a novel GPU-based rendering pipeline that
allows interactive visualization of realistic molecular
datasets comprising up to hundreds of millions of
macromolecules. The rendering pipeline is embedded into a
popular game engine and well known computer graphics
optimizations were adapted to support this type of data,
such as level-of-detail, instancing and occlusion queries.
Secondly, a new method for authoring cutaway views and
improving spatial exploration of crowded molecular
landscapes. The system relies on the use of clipping objects
that are manually placed in the scene and on visibility
equalizers that allows fine tuning of the visibility of each
species present in the scene. Agent-based modeling produces
trajectory data that can also be combined with structural
information in order to animate these landscapes. The
snapshots of the trajectories are often played in
fast-forward to shorten the length of the visualized
sequences, which also renders potentially interesting events
occurring at a higher temporal resolution invisible. The
third contribution is a solution to visualize time-lapse of
agent-based simulations that also reveals hidden information
that is only observable at higher temporal resolutions. And
finally, a new type of particle-system that utilize
quantitative models as input and generate missing spatial
information to enable the visualization of molecular
trajectories and interactions. The particle-system produces
a similar visual output as traditional agent-based modeling
tools for a much lower computational footprint and allows
interactive changing of the simulation parameters, which was
not achievable with previous methods.",
month = oct,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Institute of Computer Graphics and Algorithms, Vienna
University of Technology ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2016/LeMuzic_2016_PhD/",
}

 image
image PhD-Thesis
PhD-Thesis
