Information
- Publication Type: Conference Paper
- Workgroup(s)/Project(s):
- Date: 2016
- Location: Pilsen, Czech Republic
- Lecturer: Mathieu Le Muzic
- Booktitle: Proceedings of WSCG
Abstract
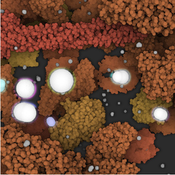
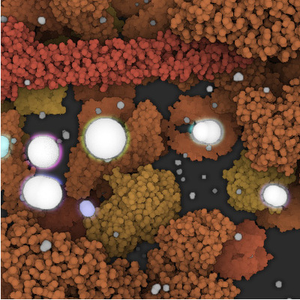
The molecular knowledge about complex biochemical reaction networks in biotechnology is crucial and has received a lot of attention lately. As a consequence, multiple visualization programs have been already developed to illustrate the anatomy of a cell. However, since a real cell performs millions of reactions every second to sustain live, it is necessary to move from anatomical to physiological illustrations to communicate knowledge about the behavior of a cell more accurately. In this thesis I propose a reaction system including a collision detection algorithm, which is able to work at the level of single atoms, to enable precise simulation of molecular interactions. To visually explain molecular activities during the simulation process, a real-time glow effect in combination with a clipping object have been implemented. Since intracellular processes are performed with a set of chemical transformations, a hierarchical structure is used to illustrate the impact of one reaction on the entire simulation. The CellPathway system integrates acceleration techniques to render large datasets containing millions of atoms in real-time, while the reaction system is processed directly on the GPU to enable simulation with more than 1000 molecules. Furthermore, a graphical user interface has been implemented to allow the user to control parameters during simulation interactively.Additional Files and Images
Weblinks
No further information available.BibTeX
@inproceedings{Reisacher2016,
title = "CellPathway: A Simulation Tool for Illustrative
Visualization of Biochemical Networks",
author = "Matthias Reisacher and Mathieu Le Muzic and Ivan Viola",
year = "2016",
abstract = "The molecular knowledge about complex biochemical reaction
networks in biotechnology is crucial and has received a lot
of attention lately. As a consequence, multiple
visualization programs have been already developed to
illustrate the anatomy of a cell. However, since a real cell
performs millions of reactions every second to sustain live,
it is necessary to move from anatomical to physiological
illustrations to communicate knowledge about the behavior of
a cell more accurately. In this thesis I propose a reaction
system including a collision detection algorithm, which is
able to work at the level of single atoms, to enable precise
simulation of molecular interactions. To visually explain
molecular activities during the simulation process, a
real-time glow effect in combination with a clipping object
have been implemented. Since intracellular processes are
performed with a set of chemical transformations, a
hierarchical structure is used to illustrate the impact of
one reaction on the entire simulation. The CellPathway
system integrates acceleration techniques to render large
datasets containing millions of atoms in real-time, while
the reaction system is processed directly on the GPU to
enable simulation with more than 1000 molecules.
Furthermore, a graphical user interface has been implemented
to allow the user to control parameters during simulation
interactively.",
location = "Pilsen, Czech Republic",
booktitle = "Proceedings of WSCG",
URL = "https://www.cg.tuwien.ac.at/research/publications/2016/Reisacher2016/",
}

 paper
paper