Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s):
- Date: 2016
- Journal: Computer Graphics Forum
- Volume: 35
- Number: 3
- Location: Groningen, Netherlands
- Lecturer: Peter Mindek
- Event: EuroVis 2016
- Conference date: 6. June 2016 – 10. June 2016
- Keywords: molecular visualization, visibility, occlusion
Abstract
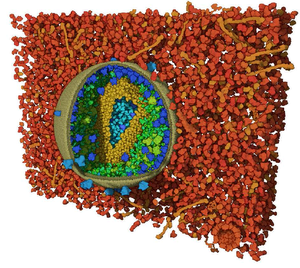
In scientific illustrations and visualization, cutaway views are often employed as an effective technique for occlusion management in densely packed scenes.We propose a novel method for authoring cutaway illustrations of mesoscopic biological models. In contrast to the existing cutaway algorithms, we take advantage of the specific nature of the biological models. These models consist of thousands of instances with a comparably smaller number of different types. Our method constitutes a two stage process. In the first step, clipping objects are placed in the scene, creating a cutaway visualization of the model. During this process, a hierarchical list of stacked bars inform the user about the instance visibility distribution of each individual molecular type in the scene. In the second step, the visibility of each molecular type is fine-tuned through these bars, which at this point act as interactive visibility equalizers. An evaluation of our technique with domain experts confirmed that our equalizer-based approach for visibility specification is valuable and effective for both, scientific and educational purposes.Additional Files and Images
Weblinks
No further information available.BibTeX
@article{lemuzic-mindek-2016-viseq,
title = "Visibility Equalizer: Cutaway Visualization of Mesoscopic
Biological Models",
author = "Mathieu Le Muzic and Peter Mindek and Johannes Sorger and
Ludovic Autin and David Goodsell and Ivan Viola",
year = "2016",
abstract = "In scientific illustrations and visualization, cutaway views
are often employed as an effective technique for occlusion
management in densely packed scenes.We propose a novel
method for authoring cutaway illustrations of mesoscopic
biological models. In contrast to the existing cutaway
algorithms, we take advantage of the specific nature of the
biological models. These models consist of thousands of
instances with a comparably smaller number of different
types. Our method constitutes a two stage process. In the
first step, clipping objects are placed in the scene,
creating a cutaway visualization of the model. During this
process, a hierarchical list of stacked bars inform the user
about the instance visibility distribution of each
individual molecular type in the scene. In the second step,
the visibility of each molecular type is fine-tuned through
these bars, which at this point act as interactive
visibility equalizers. An evaluation of our technique with
domain experts confirmed that our equalizer-based approach
for visibility specification is valuable and effective for
both, scientific and educational purposes.",
journal = "Computer Graphics Forum",
volume = "35",
number = "3",
keywords = "molecular visualization, visibility, occlusion",
URL = "https://www.cg.tuwien.ac.at/research/publications/2016/lemuzic-mindek-2016-viseq/",
}

 Paper
Paper
