Information
- Publication Type: Conference Paper
- Workgroup(s)/Project(s):
- Date: September 2016
- Organization: Eurographics
- Location: Bergen
- Editor: S. Bruckner, B. Preim, and A. Vilanova
- Booktitle: Eurographics Workshop on Visual Computing for Biology and Medicine (VCBM)
- Pages: 21 – 30
- Keywords: I.3.3 [Computer Graphics]: Picture/Image Generation-Display algorithms
Abstract
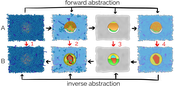
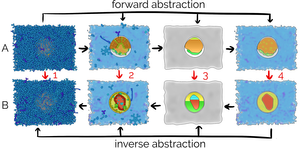
A challenging problem in biology is the incompleteness of acquired information when visualizing biological phenomena. Structural biology generates detailed models of viruses or bacteria at different development stages, while the processes that relate one stage to another are often not clear. Similarly, the entire life cycle of a biological entity might be available as a quantitative model, while only one structural model is available. If the relation between two models is specified at a lower level of detail than the actual models themselves, the two models cannot be interpolated correctly. We propose a method that deals with the visualization of incomplete data information in the developmental or evolutionary states of biological mesoscale models, such as viruses or microorganisms. The central tool in our approach is visual abstraction. Instead of directly interpolating between two models that show different states of an organism, we gradually forward transform the models into a level of visual abstraction that matches the level of detail of the modeled relation between them. At this level, the models can be interpolated without conveying false information. After the interpolation to the new state, we apply the inverse transformation to the model’'s original level of abstraction. To show the flexibility of our approach, we demonstrate our method on the basis of molecular data, in particular data of the HIV virion and the mycoplasma bacterium.Additional Files and Images
Weblinks
No further information available.BibTeX
@inproceedings{sorger-2016-fowardabstraction,
title = "Illustrative Transitions in Molecular Visualization via
Forward and Inverse Abstraction Transform",
author = "Johannes Sorger and Peter Mindek and Tobias Klein and Graham
Johnson and Ivan Viola",
year = "2016",
abstract = "A challenging problem in biology is the incompleteness of
acquired information when visualizing biological phenomena.
Structural biology generates detailed models of viruses or
bacteria at different development stages, while the
processes that relate one stage to another are often not
clear. Similarly, the entire life cycle of a biological
entity might be available as a quantitative model, while
only one structural model is available. If the relation
between two models is specified at a lower level of detail
than the actual models themselves, the two models cannot be
interpolated correctly. We propose a method that deals with
the visualization of incomplete data information in the
developmental or evolutionary states of biological mesoscale
models, such as viruses or microorganisms. The central tool
in our approach is visual abstraction. Instead of directly
interpolating between two models that show different states
of an organism, we gradually forward transform the models
into a level of visual abstraction that matches the level of
detail of the modeled relation between them. At this level,
the models can be interpolated without conveying false
information. After the interpolation to the new state, we
apply the inverse transformation to the model’'s original
level of abstraction. To show the flexibility of our
approach, we demonstrate our method on the basis of
molecular data, in particular data of the HIV virion and the
mycoplasma bacterium.",
month = sep,
organization = "Eurographics",
location = "Bergen",
editor = "S. Bruckner, B. Preim, and A. Vilanova",
booktitle = "Eurographics Workshop on Visual Computing for Biology and
Medicine (VCBM)",
pages = "21--30",
keywords = "I.3.3 [Computer Graphics]: Picture/Image Generation-Display
algorithms",
URL = "https://www.cg.tuwien.ac.at/research/publications/2016/sorger-2016-fowardabstraction/",
}

 paper
paper video
video