Information
- Publication Type: Journal Paper with Conference Talk
- Workgroup(s)/Project(s):
- Date: 2018
- Journal: IEEE Transactions on Visualization and Computer Graphics
- Lecturer: Tobias Klein
- DOI: 10.1109/TVCG.2017.2744258
Abstract
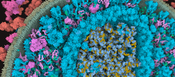
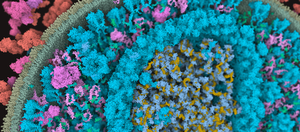
We present the first approach to integrative structural modeling of the biological mesoscale within an interactive visual environment. These complex models can comprise up to millions of molecules with defined atomic structures, locations, and interactions. Their construction has previously been attempted only within a non-visual and non-interactive environment. Our solution unites the modeling and visualization aspect, enabling interactive construction of atomic resolution mesoscale models of large portions of a cell. We present a novel set of GPU algorithms that build the basis for the rapid construction of complex biological structures. These structures consist of multiple membrane-enclosed compartments including both soluble molecules and fibrous structures. The compartments are defined using volume voxelization of triangulated meshes. For membranes, we present an extension of the Wang Tile concept that populates the bilayer with individual lipids. Soluble molecules are populated within compartments using the Halton sequence for their distribution. Fibrous structures, such as RNA or actin filaments, are created by self-avoiding random walks. Resulting overlaps of molecules are resolved by a forced-based system. Our approach opens new possibilities to the world of interactive construction of cellular compartments. We demonstrate its effectiveness by showcasing scenes of different scale and complexity that comprise blood plasma, mycoplasma, and HIV.Additional Files and Images
Weblinks
BibTeX
@article{klein_2017_IM,
title = "Instant Construction and Visualization of Crowded Biological
Environments",
author = "Tobias Klein and Ludovic Autin and Barbora Kozlikova and
David Goodsell and Arthur Olson and Eduard Gr\"{o}ller and
Ivan Viola",
year = "2018",
abstract = "We present the first approach to integrative structural
modeling of the biological mesoscale within an interactive
visual environment. These complex models can comprise up to
millions of molecules with defined atomic structures,
locations, and interactions. Their construction has
previously been attempted only within a non-visual and
non-interactive environment. Our solution unites the
modeling and visualization aspect, enabling interactive
construction of atomic resolution mesoscale models of large
portions of a cell. We present a novel set of GPU algorithms
that build the basis for the rapid construction of complex
biological structures. These structures consist of multiple
membrane-enclosed compartments including both soluble
molecules and fibrous structures. The compartments are
defined using volume voxelization of triangulated meshes.
For membranes, we present an extension of the Wang Tile
concept that populates the bilayer with individual lipids.
Soluble molecules are populated within compartments using
the Halton sequence for their distribution. Fibrous
structures, such as RNA or actin filaments, are created by
self-avoiding random walks. Resulting overlaps of molecules
are resolved by a forced-based system. Our approach opens
new possibilities to the world of interactive construction
of cellular compartments. We demonstrate its effectiveness
by showcasing scenes of different scale and complexity that
comprise blood plasma, mycoplasma, and HIV.",
journal = "IEEE Transactions on Visualization and Computer Graphics",
doi = "10.1109/TVCG.2017.2744258",
URL = "https://www.cg.tuwien.ac.at/research/publications/2018/klein_2017_IM/",
}

 paper
paper