Information
- Publication Type: Bachelor Thesis
- Workgroup(s)/Project(s):
- Date: April 2019
- Date (Start): 10. October 2018
- Date (End): 18. April 2019
- Matrikelnummer: 01307685
- First Supervisor: Eduard Gröller
Abstract
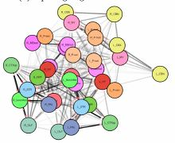
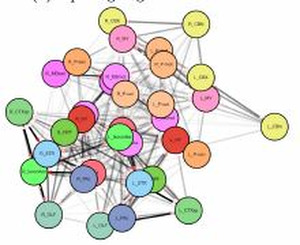
The visualization of brain networks today offers a variety of different tools and approaches. Representations in 2D such as connectograms, connectivity matrices, and node-link diagrams are common but an abstract visualization of the network without any anatomical context. Visualizations tools show anatomical context in 2D but adjust it especially for a certain species as for example the fruit fly’s brain. This project presents a tool for data-driven brain network visualization using the open-source graph library Cytoscape.js to avoid hard coded spatial constraints. The goal of the project was to find a layout algorithm that resembles the anatomical structure of the brain visualized without any hard coded constraints. After testing the layouts, they have been evaluated on different properties like symmetry, node overlapping, and anatomical resemblence. Additionally, we conducted an open discussion with collaborators of the Research Institute of Molecular Pathology (IMP) in Vienna and present the results.Additional Files and Images
Weblinks
No further information available.BibTeX
@bachelorsthesis{Rippberger_2019,
title = "Data-Driven Anatomical Layouting of Brain Network Graphs",
author = "Gwendolyn Rippberger",
year = "2019",
abstract = "The visualization of brain networks today offers a variety
of different tools and approaches. Representations in 2D
such as connectograms, connectivity matrices, and node-link
diagrams are common but an abstract visualization of the
network without any anatomical context. Visualizations tools
show anatomical context in 2D but adjust it especially for a
certain species as for example the fruit fly’s brain. This
project presents a tool for data-driven brain network
visualization using the open-source graph library
Cytoscape.js to avoid hard coded spatial constraints. The
goal of the project was to find a layout algorithm that
resembles the anatomical structure of the brain visualized
without any hard coded constraints. After testing the
layouts, they have been evaluated on different properties
like symmetry, node overlapping, and anatomical resemblence.
Additionally, we conducted an open discussion with
collaborators of the Research Institute of Molecular
Pathology (IMP) in Vienna and present the results.",
month = apr,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Research Unit of Computer Graphics, Institute of Visual
Computing and Human-Centered Technology, Faculty of
Informatics, TU Wien ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/Rippberger_2019/",
}
 Bachelor Thesis
Bachelor Thesis image
image