Information
- Publication Type: PhD-Thesis
- Workgroup(s)/Project(s):
- Date: June 2019
- Date (Start): 2014
- Date (End): 2019
- TU Wien Library:
- Second Supervisor: Katja Bühler

- Open Access: yes
- 1st Reviewer: Anna Vilanova i Bartroli

- 2nd Reviewer: Torsten Kuhlen
- Rigorosum: 3. September 2019
- First Supervisor: Eduard Gröller

Abstract
Advances in neuro-imaging have allowed big brain initiatives and consortia to create vast resources of brain data that can be mined for insights into mental processes and biological principles. Research in this area does not only relate to mind and consciousness, but also to the understanding of many neurological disorders, such as Alzheimer’s disease, autism, and anxiety. Exploring the relationships between genes, brain circuitry, and behavior is therefore a key element in research that requires the joint analysis of a heterogeneous set of spatial brain data, including 3D imaging data, anatomical data, and brain networks at varying scales, resolutions, and modalities. Due to high-throughput imaging platforms, this data’s size and complexity goes beyond the state-of-the-art by several orders of magnitude. Current analytical workflows involve time-consuming manual data aggregation and extensive computational analysis in script-based toolboxes. Visual analytics methods for exploring big brain data can support neuroscientists in this process, so they can focus on understanding the data rather than handling it.
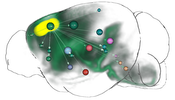
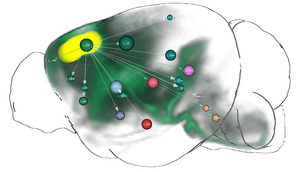
In this thesis, several contributions that target this problem are presented. The first contribution is a computational method that fuses genetic information with spatial gene expression data and connectivity data to predict functional neuroanatomical maps. These maps indicate, which brain areas might be related to a specific function or behavior. The approach has been applied to predict yet unknown functional neuroanatomy underlying multigeneic behavioral traits identified in genetic association studies and has demonstrated that rather than being randomly distributed throughout the brain, functionally-related gene sets accumulate in specific networks. The second contribution is the creation of a data structure that enables the interactive exploration of big brain network data with billions of edges. By utilizing the resulting hierarchical and spatial organization of the data, this approach allows neuroscientists on-demand queries of incoming/outgoing connections of arbitrary regions of interest on different anatomical scales. These queries would otherwise exceed the limits of current consumer level PCs. The data structure is used in the third contribution, a novel web-based framework to explore neurobiological imaging and connectivity data of different types, modalities, and scale. It employs a query-based interaction scheme to retrieve 3D spatial gene expressions and various types of connectivity to enable an interactive dissection of networks in real-time with respect to their genetic composition. The data is related to a hierarchical organization of common anatomical atlases that enables neuroscientists to compare multimodal networks on different scales in their anatomical context. Furthermore, the framework is designed to facilitate collaborative work with shareable comprehensive workflows on the web.
As a result, the approaches presented in this thesis may assist neuroscientists to refine their understanding of the functional organization of the brain beyond simple anatomical domains and expand their knowledge about how our genes affect our mind.
Additional Files and Images
Additional images and videos
Additional files
Weblinks
No further information available.
BibTeX
@phdthesis{ganglberger2019,
title = "From Neurons to Behavior: Visual Analytics Methods for
Heterogeneous Spatial Big Brain Data ",
author = "Florian Ganglberger",
year = "2019",
abstract = "Advances in neuro-imaging have allowed big brain initiatives
and consortia to create vast resources of brain data that
can be mined for insights into mental processes and
biological principles. Research in this area does not only
relate to mind and consciousness, but also to the
understanding of many neurological disorders, such as
Alzheimer’s disease, autism, and anxiety. Exploring the
relationships between genes, brain circuitry, and behavior
is therefore a key element in research that requires the
joint analysis of a heterogeneous set of spatial brain data,
including 3D imaging data, anatomical data, and brain
networks at varying scales, resolutions, and modalities. Due
to high-throughput imaging platforms, this data’s size and
complexity goes beyond the state-of-the-art by several
orders of magnitude. Current analytical workflows involve
time-consuming manual data aggregation and extensive
computational analysis in script-based toolboxes. Visual
analytics methods for exploring big brain data can support
neuroscientists in this process, so they can focus on
understanding the data rather than handling it. In this
thesis, several contributions that target this problem are
presented. The first contribution is a computational method
that fuses genetic information with spatial gene expression
data and connectivity data to predict functional
neuroanatomical maps. These maps indicate, which brain areas
might be related to a specific function or behavior. The
approach has been applied to predict yet unknown functional
neuroanatomy underlying multigeneic behavioral traits
identified in genetic association studies and has
demonstrated that rather than being randomly distributed
throughout the brain, functionally-related gene sets
accumulate in specific networks. The second contribution is
the creation of a data structure that enables the
interactive exploration of big brain network data with
billions of edges. By utilizing the resulting hierarchical
and spatial organization of the data, this approach allows
neuroscientists on-demand queries of incoming/outgoing
connections of arbitrary regions of interest on different
anatomical scales. These queries would otherwise exceed the
limits of current consumer level PCs. The data structure is
used in the third contribution, a novel web-based framework
to explore neurobiological imaging and connectivity data of
different types, modalities, and scale. It employs a
query-based interaction scheme to retrieve 3D spatial gene
expressions and various types of connectivity to enable an
interactive dissection of networks in real-time with respect
to their genetic composition. The data is related to a
hierarchical organization of common anatomical atlases that
enables neuroscientists to compare multimodal networks on
different scales in their anatomical context. Furthermore,
the framework is designed to facilitate collaborative work
with shareable comprehensive workflows on the web. As a
result, the approaches presented in this thesis may assist
neuroscientists to refine their understanding of the
functional organization of the brain beyond simple
anatomical domains and expand their knowledge about how our
genes affect our mind. ",
month = jun,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Research Unit of Computer Graphics, Institute of Visual
Computing and Human-Centered Technology, Faculty of
Informatics, TU Wien ",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/ganglberger2019/",
}

 image
image PhD Thesis
PhD Thesis