Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: March 2019
- Date (Start): 2018
- Date (End): 5. March 2019
- TU Wien Library:
- First Supervisor: Ivan Viola
Abstract
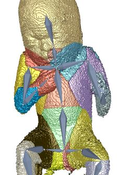
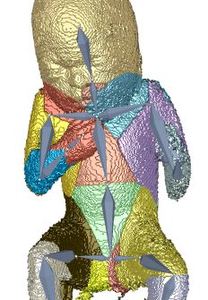
Three dimensional ultrasound images are commonly used in prenatal screening. The acquisition delivers detailed information about the skin as well as the inner organs of the fetus. Prenatal screenings in terms of growth analysis are very important to support a healthy development of the fetus. The analysis of this data involves viewing of two dimensional (2D) slices in order to take measurements or calculate the volume and weight of the fetus. These steps involve manual investigation and are dependent on the skills of the person who performs them. These measurements and calculations are very important to analyze the development of the fetus and for the birth preparation. Ultrasound imaging is a˙ected by artifacts like speckles, noise and also of structures obstructing the regions of interest. These artifacts occur because the imaging technique is using sound waves and their echo to create images. 2D slices as used as basis for the measurement of the fetus therefore might not be the best solution. Analyzing the data in a three dimensional (3D) way would enable the viewer to have a better overview and to better distinguish between artifacts and the real data of the fetus. The growth of a fetus can be analysed by comparing standardized measurements like the crown foot length, the femur length or the derived head circumference as well as the abdominal circumference. Standardization is well known in many fields of medicine and is used to enable compa-rability between investigations of the same patient or between patients. Therefore we introduce a standardized way of analyzing 3D ultrasound images of fetuses. Bringing the fetus in a standardized position would enable automatized measurements by the machine and there could also be new measurements applied like the volume of specific body parts. A standardized pose would also provide possibilities to compare the re-sults of di˙erent measurements of one fetus as well as the measurements of di˙erent fetuses. The novel method consists of six steps, namely the loading of the data, the preprocessing, the rigging of the model, the weighting of the data, the actual transformation called the "Vitruvian Baby" and at the end the analysis of the result. We tried to automatize the workflow as far as possible resulting in some manual tasks and some automatic ones. The loading of the data works with standard medical image formats and the preprocessing involves some interaction in order to get rid of the ultrasound induced artifacts. Transforming data into a specific position is a complex task which might involve a manual processing steps. In the method presented in this work one step of the transformation namely the rigging of the model, where a skeleton is placed in the data, is performed manually. The weighting as well as the transformation although are performed completely automatically resulting in a T-pose representation of the data. We analysed the performance of our novel approach in several ways. We first use a phantom model which has been used as a reference already presented in a T-pose. After using seven di˙erent fetus poses of the model as input the result was an average of 79,02%voxel overlapping between the output of the method and the goal T-pose. When having a look at the similarity of the finger to finger span and the head to toe measurement we considered a value of 91,08% and 94,05% in average. The time needed for the most complex manual task was in average seven minutes. After using a phantom model of a man, we also assessed the performance of the method using a computer model of a fetus and a phantom model of a 3D ultrasound investigation. The results also look very promising.Additional Files and Images
Weblinks
BibTeX
@mastersthesis{moerth-2018-tpose,
title = "Interactive Reformation of Fetal Ultrasound Data to a
T-Position",
author = "Eric M\"{o}rth",
year = "2019",
abstract = "Three dimensional ultrasound images are commonly used in
prenatal screening. The acquisition delivers detailed
information about the skin as well as the inner organs of
the fetus. Prenatal screenings in terms of growth analysis
are very important to support a healthy development of the
fetus. The analysis of this data involves viewing of two
dimensional (2D) slices in order to take measurements or
calculate the volume and weight of the fetus. These steps
involve manual investigation and are dependent on the skills
of the person who performs them. These measurements and
calculations are very important to analyze the development
of the fetus and for the birth preparation. Ultrasound
imaging is a˙ected by artifacts like speckles, noise and
also of structures obstructing the regions of interest.
These artifacts occur because the imaging technique is using
sound waves and their echo to create images. 2D slices as
used as basis for the measurement of the fetus therefore
might not be the best solution. Analyzing the data in a
three dimensional (3D) way would enable the viewer to have a
better overview and to better distinguish between artifacts
and the real data of the fetus. The growth of a fetus can be
analysed by comparing standardized measurements like the
crown foot length, the femur length or the derived head
circumference as well as the abdominal circumference.
Standardization is well known in many fields of medicine and
is used to enable compa-rability between investigations of
the same patient or between patients. Therefore we introduce
a standardized way of analyzing 3D ultrasound images of
fetuses. Bringing the fetus in a standardized position would
enable automatized measurements by the machine and there
could also be new measurements applied like the volume of
specific body parts. A standardized pose would also provide
possibilities to compare the re-sults of di˙erent
measurements of one fetus as well as the measurements of
di˙erent fetuses. The novel method consists of six steps,
namely the loading of the data, the preprocessing, the
rigging of the model, the weighting of the data, the actual
transformation called the "Vitruvian Baby" and at the end
the analysis of the result. We tried to automatize the
workflow as far as possible resulting in some manual tasks
and some automatic ones. The loading of the data works with
standard medical image formats and the preprocessing
involves some interaction in order to get rid of the
ultrasound induced artifacts. Transforming data into a
specific position is a complex task which might involve a
manual processing steps. In the method presented in this
work one step of the transformation namely the rigging of
the model, where a skeleton is placed in the data, is
performed manually. The weighting as well as the
transformation although are performed completely
automatically resulting in a T-pose representation of the
data. We analysed the performance of our novel approach in
several ways. We first use a phantom model which has been
used as a reference already presented in a T-pose. After
using seven di˙erent fetus poses of the model as input the
result was an average of 79,02%voxel overlapping between the
output of the method and the goal T-pose. When having a look
at the similarity of the finger to finger span and the head
to toe measurement we considered a value of 91,08% and
94,05% in average. The time needed for the most complex
manual task was in average seven minutes. After using a
phantom model of a man, we also assessed the performance of
the method using a computer model of a fetus and a phantom
model of a 3D ultrasound investigation. The results also
look very promising.",
month = mar,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Research Unit of Computer Graphics, Institute of Visual
Computing and Human-Centered Technology, Faculty of
Informatics, TU Wien",
URL = "https://www.cg.tuwien.ac.at/research/publications/2019/moerth-2018-tpose/",
}

 image
image Master Thesis
Master Thesis