Information
- Publication Type: Master Thesis
- Workgroup(s)/Project(s):
- Date: February 2020
- Date (Start): 20. September 2019
- Date (End): 27. February 2020
- TU Wien Library:
- Second Supervisor: Katja Bühler
- Diploma Examination: May 2020
- Open Access: yes
- First Supervisor: Eduard Gröller
Abstract
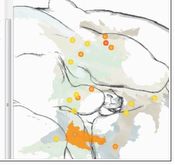
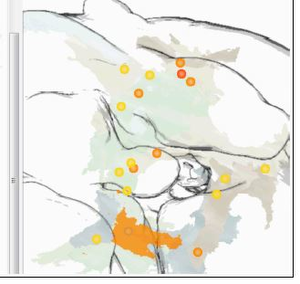
In their studies of the brain of the common fruit fly Drosophila melanogaster, neuro-biologists investigate neural connectivity with the goal to discover how complex be-haviour is generated. Gaining information on potential connectivity between neurons is an essential step in their workflow. This thesis presents a way to compute and visualise such potential connectivity information from segmented neurons. It shortens the tedious month-long search for potential connectivity down to a few minutes. Overlaps of arborisations of two or more neurons indicate a potential anatomical connection, and thus a potential functional connection. The computation of this data starts from neuron meshes. The meshes—segmented from confocal light-microscopy images of the fruit fly—are intersected to find overlapping areas, i.e. areas of potential anatomical connectivity. This information can then help to discover actual functional connectivity in a neural circuit. Analysing higher order overlaps, i.e. intersections of more than two arborisations of segmented neuron data in the same location, poses new challenges. The visualisation in 2D sections or 3D is impeded by visual clutter and occlusion. Computation of relevant volumetric information becomes difficult for higher order overlaps, because the number of possible overlaps increases exponentially with the number of arborisations. This makes the pre-computation for all possible combinations infeasible. Previous tools have thus been restricted to the quantification and visualisation of pairwise overlaps. The thesis presents a novel solution addressing these issues for higher order over-laps. A novel abstracting design is coupled with a modern approach for on-demand GPU computations. Our tool calculates for the first time volumetric information of higher order overlaps on the GPU using A-buffers. The thesis addresses the visual com-plexity of the data with the implementation of an innovative novel design created by the graphics designer Judith Moosburner. We realised this design using non-photorealistic rendering techniques and perspicuous user interfaces, including interactive glyphs and linked views on quantitative overlap information. To complement the neuroscientists’ workflows the resulting interactive tool has been integrated into BrainGazer, a software tool for advanced visualisation and exploration of neural images and circuit data. Qual-itative evaluation with neuroscientists and non-expert users demonstrated the utility and usability of the tool.Additional Files and Images
Weblinks
No further information available.BibTeX
@mastersthesis{Swoboda2020,
title = "Visualisation and Interaction Techniques for the Exploration
of the Fruit Fly’s Neural Structure",
author = "Nicolas Swoboda",
year = "2020",
abstract = "In their studies of the brain of the common fruit fly
Drosophila melanogaster, neuro-biologists investigate neural
connectivity with the goal to discover how complex
be-haviour is generated. Gaining information on potential
connectivity between neurons is an essential step in their
workflow. This thesis presents a way to compute and
visualise such potential connectivity information from
segmented neurons. It shortens the tedious month-long search
for potential connectivity down to a few minutes. Overlaps
of arborisations of two or more neurons indicate a potential
anatomical connection, and thus a potential functional
connection. The computation of this data starts from neuron
meshes. The meshes—segmented from confocal
light-microscopy images of the fruit fly—are intersected
to find overlapping areas, i.e. areas of potential
anatomical connectivity. This information can then help to
discover actual functional connectivity in a neural circuit.
Analysing higher order overlaps, i.e. intersections of more
than two arborisations of segmented neuron data in the same
location, poses new challenges. The visualisation in 2D
sections or 3D is impeded by visual clutter and occlusion.
Computation of relevant volumetric information becomes
difficult for higher order overlaps, because the number of
possible overlaps increases exponentially with the number of
arborisations. This makes the pre-computation for all
possible combinations infeasible. Previous tools have thus
been restricted to the quantification and visualisation of
pairwise overlaps. The thesis presents a novel solution
addressing these issues for higher order over-laps. A novel
abstracting design is coupled with a modern approach for
on-demand GPU computations. Our tool calculates for the
first time volumetric information of higher order overlaps
on the GPU using A-buffers. The thesis addresses the visual
com-plexity of the data with the implementation of an
innovative novel design created by the graphics designer
Judith Moosburner. We realised this design using
non-photorealistic rendering techniques and perspicuous user
interfaces, including interactive glyphs and linked views on
quantitative overlap information. To complement the
neuroscientists’ workflows the resulting interactive tool
has been integrated into BrainGazer, a software tool for
advanced visualisation and exploration of neural images and
circuit data. Qual-itative evaluation with neuroscientists
and non-expert users demonstrated the utility and usability
of the tool.",
month = feb,
address = "Favoritenstrasse 9-11/E193-02, A-1040 Vienna, Austria",
school = "Research Unit of Computer Graphics, Institute of Visual
Computing and Human-Centered Technology, Faculty of
Informatics, TU Wien",
URL = "https://www.cg.tuwien.ac.at/research/publications/2020/Swoboda2020/",
}
 image
image Master Thesis
Master Thesis Poster
Poster